Common Contaminants
The quality of sequence data is directly related to the quality of the template. Several companies market plasmid purification kits. Most of these kits will work for DNA sequencing. You may need to try a couple of kits to find the method that works best in your lab. The Genomics Technology Core can provide information on plasmid preparation and purification of PCR products for sequencing.
Excess Salt
Salts can be introduced in a sample through poor elution from a column or a poor wash step during the precipitation method. Excess salts severely inhibit polymerase activity and often is the culprit for sample to sample variability. Figure 1 illustrates the effect of increased salt concentration with the addition of NaCl to the reaction. Figure 1a is of pGEM control sequence from a reaction performed with no salts added to the reaction. The peaks are well resolved with good signal strength. The sequence was 98.5% accurate in the first 861 bases. Figure 1b contains sequence data of a pGEM control reaction in the presence of 20mM NaCl. The peaks are well resolved but signal strength is decreased. Read length and sequence quality is reduced with sequence being 98.5% accurate in the first 695 bases. Figure 1c contains sequence data of a pGEM control reaction in the presence of 40mM NaCl. Data shows a much more dramatic reduction in signal strength. Incorrect base calls are observed. Sequence quality is reduced with sequence 98.5% accurate in the first 640 bases. The addition of the 40mM NaCl resulted in a loss of 220 bases from the read length of the control sequence. Increased amounts of salt in the reaction beyond the 40mM concentration would eventually inhibit the reaction entirely.
(a) pGEM Control
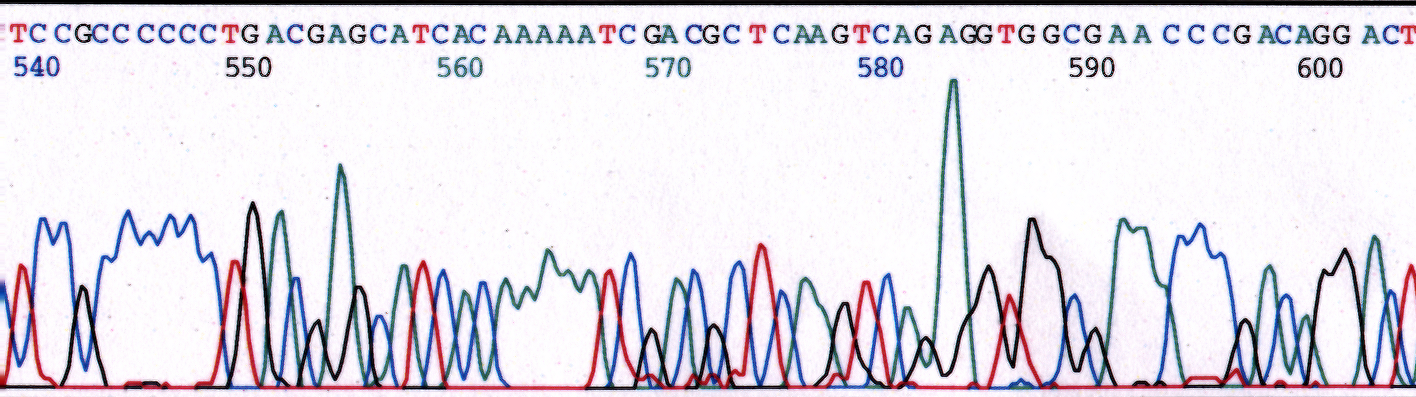
(b) 20mM NaCl
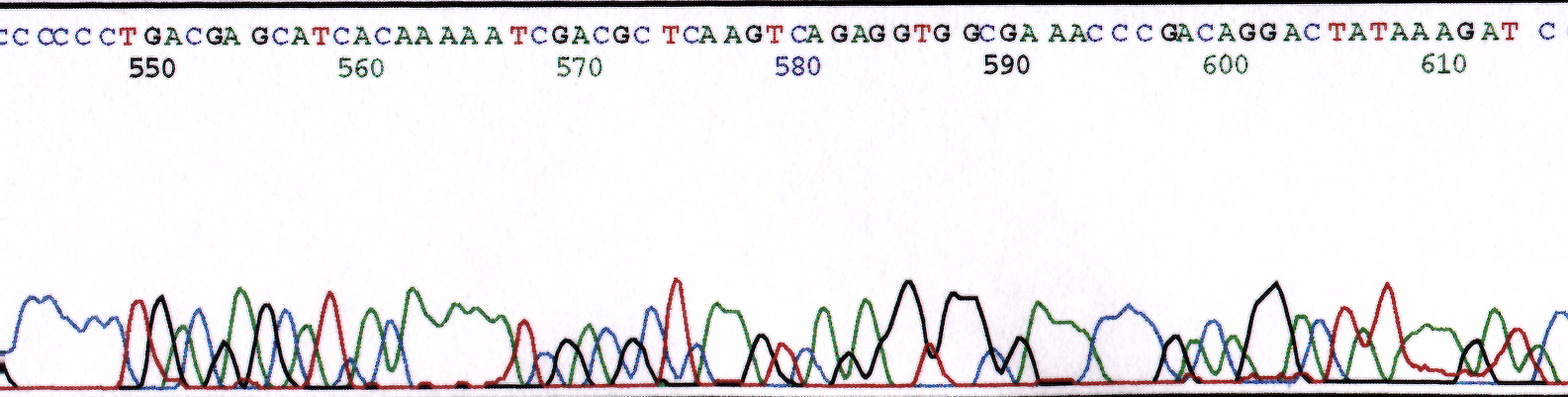
(c) 40mM NaCl
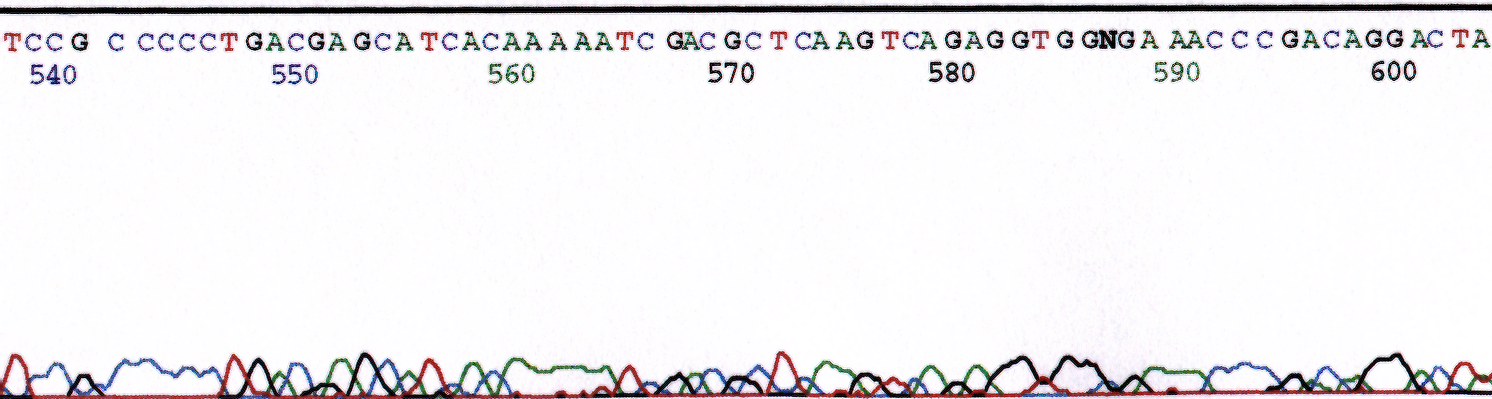
Figure 1. Chromatograms of sequence reactions containing pGEM plasmid (800ng) and M13 Forward (5uM). (a) No NaCl added to reaction (b) 20mM NaCl added to reaction (c) 40mM NaCl added to reaction
EDTA
EDTA is a potent inhibitor of the sequence reaction. Template in buffer containing 1 mM EDTA is acceptable. The final concentration in the sequencing reaction (20ul) will be 0.5 mM or less depending on the volume of template added. A final EDTA concentration greater than 1 mM will inhibit the reaction by chelating critical cations. Figure 2 illustrates the inhibiting activity of EDTA.
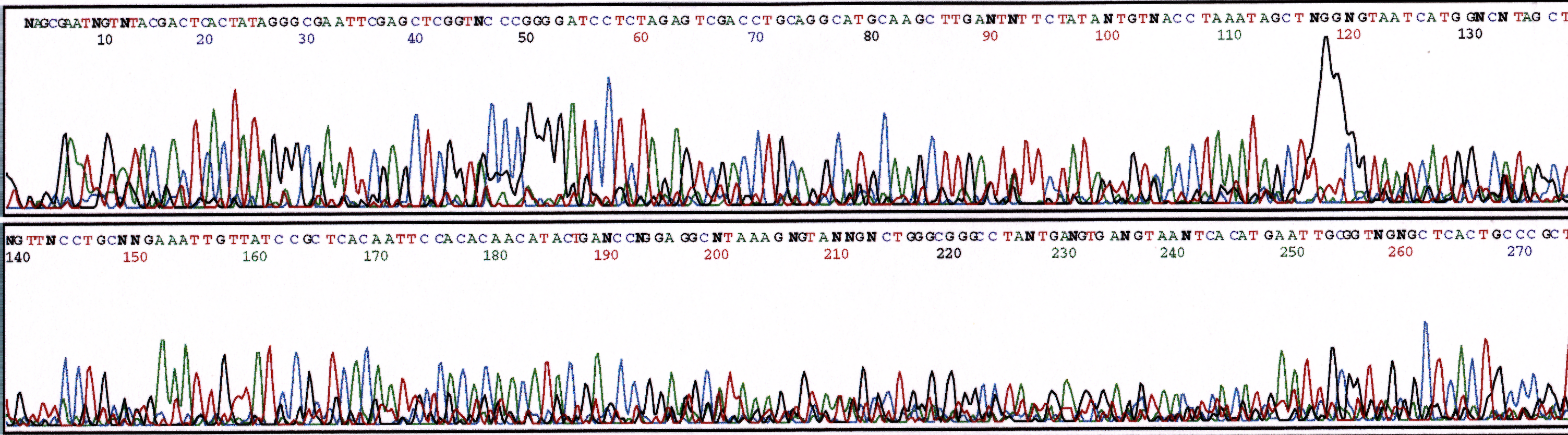
Figure 2. Chromatogram of sequence reaction containing a final volume of 2.5mM EDTA. Sequence reaction performed with pGEM plasmid (800ng) and M13 Forward (5uM).
Ethanol
The presence of ethanol will inhibit polymerase activity. Ethanol contamination most commonly occurs to samples that have been precipitated with ethanol and residual ethanol remaining from an incomplete drying step. Figure 7 displays a set of reaction results that have been spiked with increasing amounts of ethanol. Small amounts of ethanol can be tolerated in the sequence reaction. The polymerase is almost entirely inhibited, however, with a final volume of 10% ethanol as shown in Figure 3c.
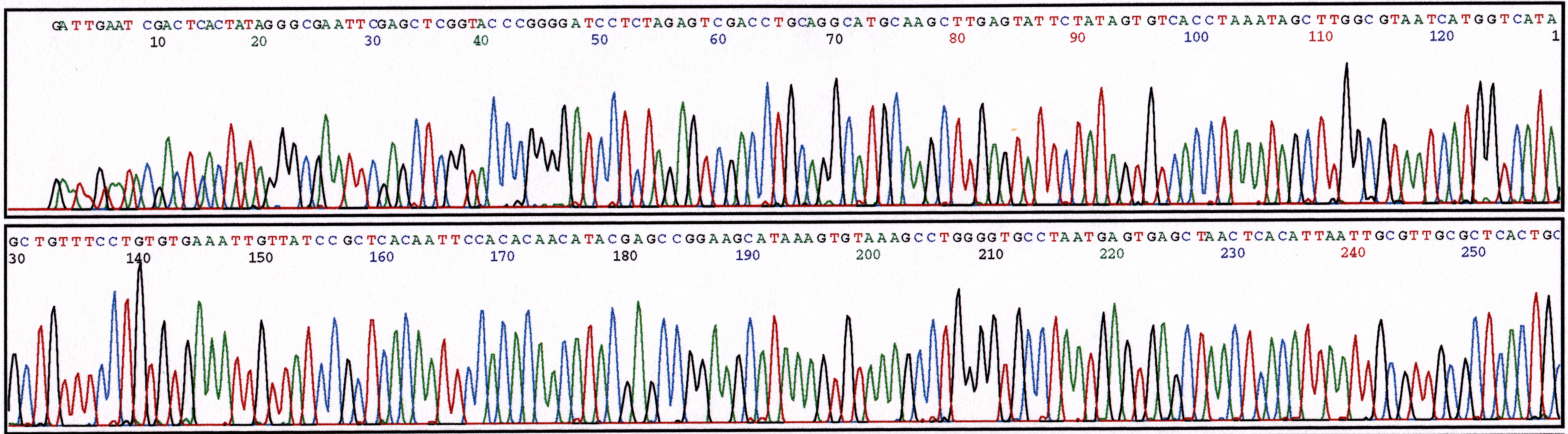
(a) 2.5% Ethanol
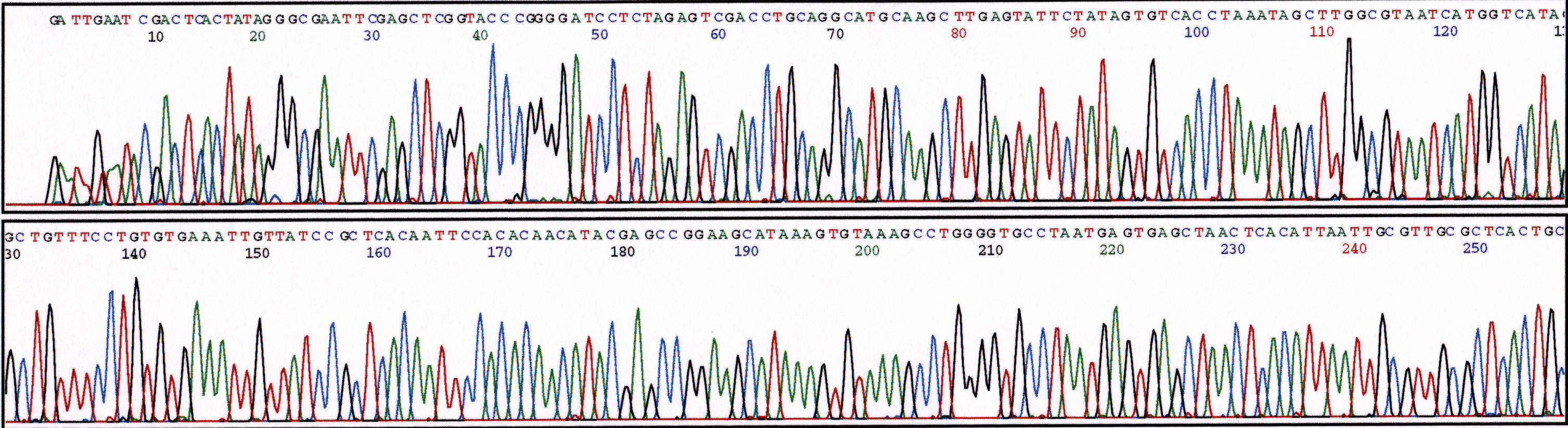
(b) 5% Ethanol
(c) 10% Ethanol
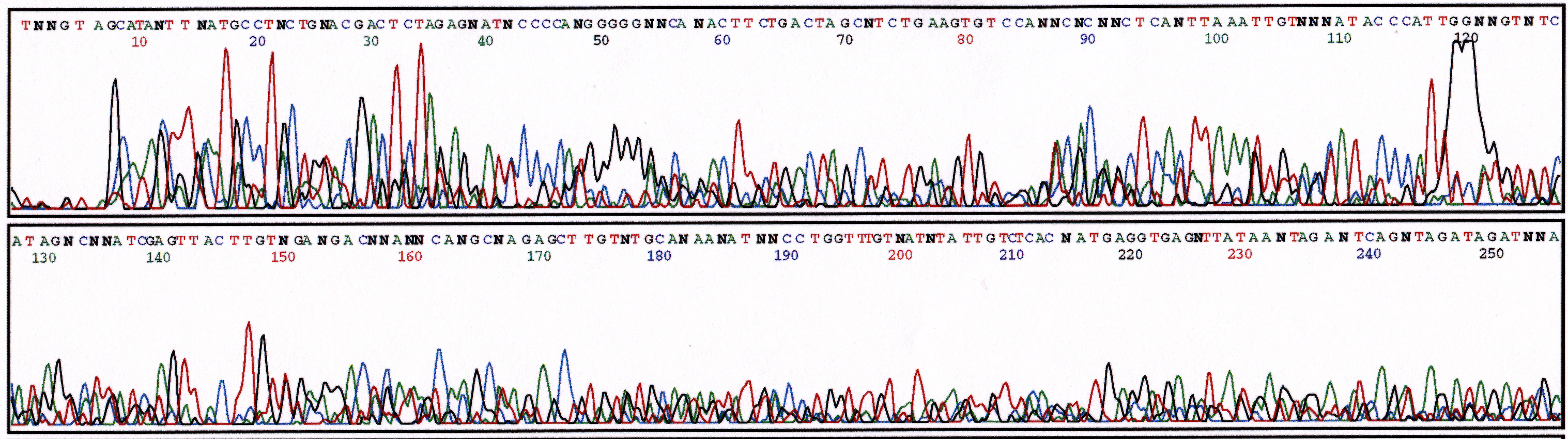
Figure 3. Series of sequencing reactions with increased amounts of ethanol. Sequence reactions performed with pGEM plasmid (800ng) and M13 Forward primer (5 uM). (a) 2.5% ethanol (b) 5% ethanol (c) 10% ethanol.
Cellular Constituents
Poor technique during template preparation will contaminate the sample with cellular constituents that will inhibit polymerase activity giving poor results.
Nucleases
Template degradation due to nuclease contamination will result in poor sequence data.
