Common Editing Problems
Once the quality of the sequence data is determined to be satisfactory, the sequence will need to be edited. Base miscalls by the analysis software are common and should be expected. The following are examples of these and other more common base calling problems.
Insertions
The insertion of an extra base in the sequence is common near the end of a sequence run. As the resolution deteriorates the peaks broaden. The analysis software uses a set value called base spacing to locate peaks in the chromatogram. The base spacing is optimal for the middle region of collected data where resolution is best, but not optimal for the end, or beginning, where resolution is poor. The broad peaks at the end of a run can lead to a single peak being assigned as two bases by the analysis software. The chromatogram in Figure 1 illustrates base insertions that occur between bases 640 and 650. The A at position 645 is an extra base assigned to the A peak. The same is true for the T at position 648. (Also notice that the G directly under the A at position 645 has been missed! Deletions are discussed next.)
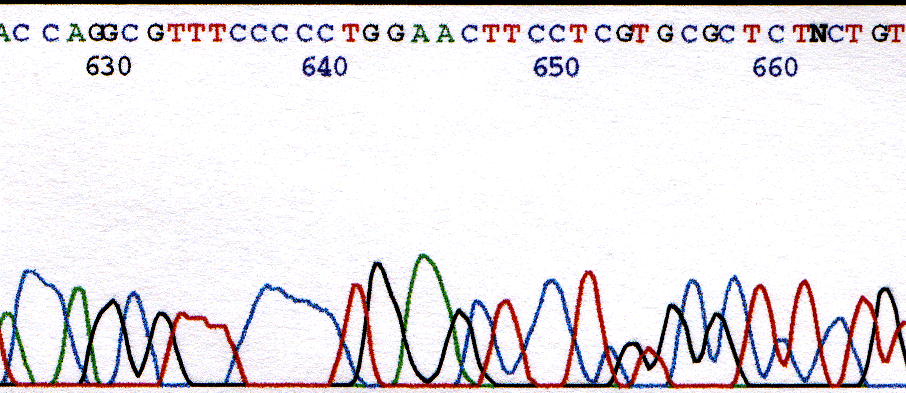
Figure 1. Chromatogram illustrating insertion of extra bases at position 645 and 648.
Deletions
The exclusion of a base is most common near the beginning of a sequence run, but can be found throughout the entire sequence. Resolution is poor in the beginning of the sequence with peaks sometimes overlapping. Due to the analysis software looking for bases at set intervals, a peak can be missed. Observe the missing A after the G at base 14 in Figure 2. There are two distinct green (A) peaks but the analysis software has only called one base.
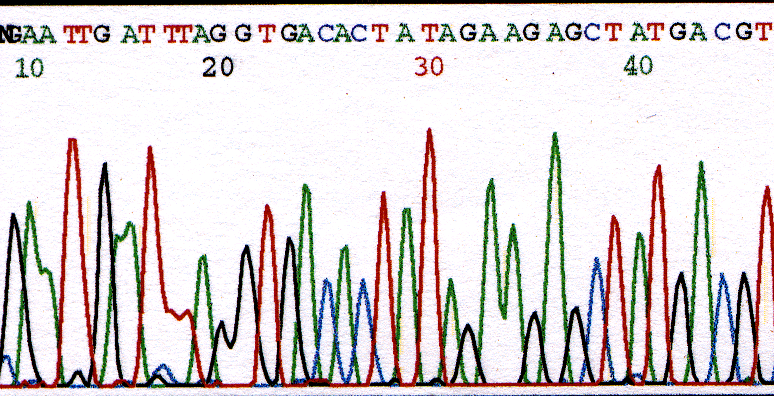
Figure 2. Chromatogram illustrating overlooked base at position 14.
Weak G's after A's
A common base miscall is a G that follows an A. The rate of incorporation of G's after A's by the enzyme is low. Compare the signal intensity of the G at base 372 and 375 with that G at base 391 in Figure 3.
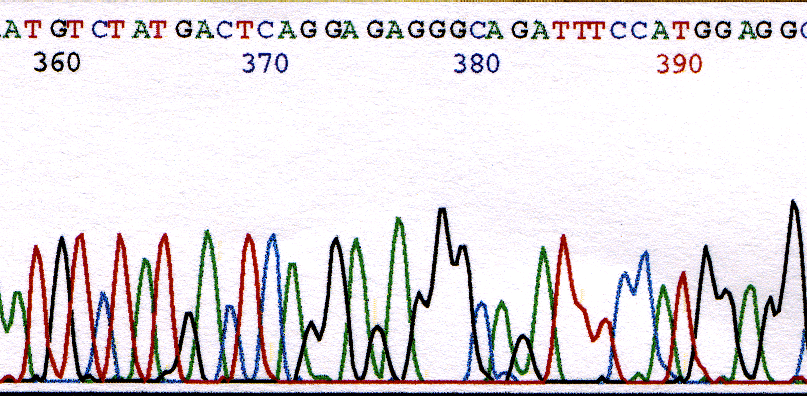
Figure 3. Chromatogram illustrating the weak G's that follow A's at position 372 and 375.
The G's after A's in Figure 15 are weak but not miscalled by the software. The G, however, can be so weak that the software is unable to assign the base. The G at position 389 and 391 in Figure 4 were assigned as an N because they were too weak causing them to be hidden by the large A peaks.
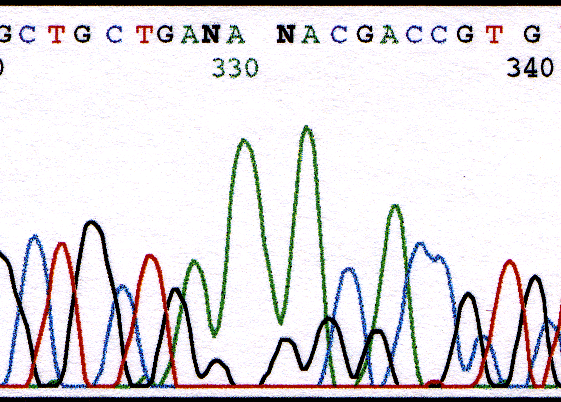
Figure 4. Chromatogram illustrating the weak G's that follow A's at position 329 and 331 which are too weak to be called.
