 The Genomics Technology Core applies high-throughput DNA sequencing to the study of complex bacterial and fungal communities. Several methods are employed:
The Genomics Technology Core applies high-throughput DNA sequencing to the study of complex bacterial and fungal communities. Several methods are employed:
- Targeted sequencing of the ITS regions for characterization of fungal communities
- Targeted sequencing of hyper-variable regions of the 16S gene to characterize bacterial communities.
- Metatranscriptomics to evaluate expressed genes.
- Whole genome shotgun sequencing
Core staff are available to assist with experimental design and to answer questions related to metagenomic approaches. Arrangements for a meeting to discuss new or existing projects can be made by sending an email to Nathan Bivens, core director.
Assistance with grant proposals and letters of support is also available by contacting the director.
Prior to sample submission and services performed, a project must be initiated with the core director and involves the following steps.
- Contact the core director to discuss project details. Several parameters are needed to quote a project and establish the scope of the work to be performed.
- Number of samples
- Organism/sample source
- Number of sequence reads per sample to be generated.
- MO code for project charges.
- The core director will prepare a Project Authorization Form (PAF) outlining the services. The form must be signed and returned before services are started.
- A Project Information Form should be completed and submitted by email prior to sample submission.
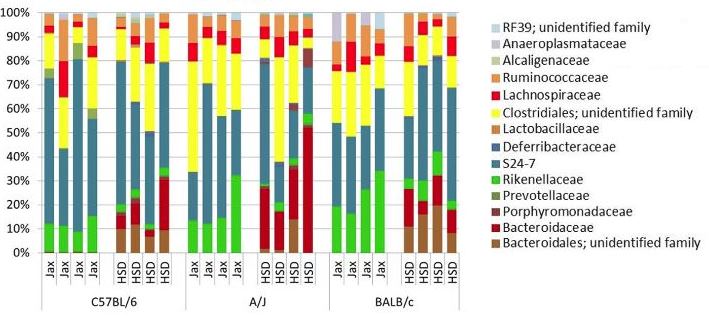
The MUGTC provides a targeted amplicon approach for evaluating and understanding complex bacterial communities. Our pipeline provides a streamlined process for the analysis of 96 samples simutaneously.
The Bioinformatics and Analytics Core can provide an analysis of the data using the Qiime pipeline, http://qiime.org/. This will provide graphical and tabular summaries of the taxa and OTUs identified and their relative proportional representations.
Standard amplicon primer sets:
| Target Region | Primer Name | Primer Sequence | Amplicon Length (nt)* | ||
| 16S V4 | U515F | 5'-GTGCCAGCMGCCGCGGTAA-3' | 430 | ||
| 806R | 5'-GGACTACHVGGGTWTCTAAT-3' | ||||
| ITS1 | ITS1-30F | 5'-GTCCCTGCCCTTTGTACACA-3' | 535 | ||
| ITS1-217R | 5'-TTTCGCTGCGTTCTTCATCG-3' | ||||
Custom amplicon primer design:
| PCR Round | Primer Name | Primer Sequence | |||
| Round 1* | Forward Primer | 5'-acactctttccctacacgacgctcttccgatct NNNNNNNNNNN-3' | |||
| Reverse Primer | 5'-gtgactggagttcagacgtgtgctcttccgatct NNNNNNNNNNN-3' | ||||
| * First round amplification to be performed by the researcher. | |||||
| Round 2 | i5 Index Primer | 5'-aatgatacggcgaccaccgagatctacac[i5]acactctttccctacacgacgctcttccgatct-3' | |||
| i7 Index Primer | 5'-caagcagaagacggcatacgagat[i7]gtgactggagttcagacgtgtgctcttccgatct-3' | ||||
Design considerations:
- Lowercase letters denote Illumina adapter sequences necessary for binding to the flow cell
- Underlined lowercase are binding sites for the Illumina sequencing primers
- [i5] and [i7] refer to dual indexing sites; 8 bases per index
- Sequence of uppercase "N's" are region specific portion of round 1 primers
General Considerations:
- Submit cells in Eppendorf DNA Lo Bind Tubes, 0.2ml (Cat. No. 022431048)
- The preferred 96-well plate for submitting targeted amplicon samples is from MedSupply Partners (877-633-7779, http://www.medsupplypartners.com/); catalog # 15-3590. The core recommends the use of adhesive sealing sheets from Thermo Scientific (cat. #AB-0558).
| Library Type | Quantity Required | Concentration | Recommended Buffer |
Special Instructions |
| DNA (genomic) | 1000 - 2000 ng | 100 - 200 ng/µl | EB buffer; 10 mM Tris-Cl (pH 8.5) | An image of 100 ng of genomic DNA following agarose gel electrophoresis and ethidium bromide staining will be provided by the researcher to demonstrate gDNA integrity. |
| Total RNA Stranded (rRNA depletion) | 500 - 1000 ng | 50 - 200 ng/µl | nuclease-free water | Provide sample source and extraction information on methods used. |
| Target amplicon (16S & ITS) | 210 ng | 3.5 ng/µlvolume = 60 µl | EB buffer; 10 mM Tris-Cl (pH 8.5) | We strongly recommend DNA be quantified by Qubit assay. |
| Custom amplicon (2-step PCR) | 60 ng | 1 ng/µlvolume = 60 µl | EB buffer; 10 mM Tris-Cl (pH 8.5) | We strongly recommend DNA be quantified by Qubit assay. |
| Library Type | Sequence Depth* | Sequence Type | Sequence Read | Cycle Number | |
| 16S V4 Amplicon | 100,000 reads/sample | Paired-end, unique, dual indexing |
|
|
|
| ITS Amplicon | 100,000 reads/sample | Paired-end, unique, dual indexing |
|
|
|
| Metatranscriptome | 20 million reads/sample | Paired-end, unique, dual indexing |
|
|
|
* minimum read count recommended; increased sequencing depth may be desired for some studies
| Library Prep Type | Fee |
|
|
| NovaSeq 6000 | Fee |
|
|
| MiSeq | Fee |
|
|
