Multiple Templates
A sequence reaction that yields more than one sequence product results in multiple peaks at each nucleotide position. Figure 1 displays the appearance of two sequences superimposed over the other in a single chromatogram. Observe that the peaks are well resolved but give the same ambiguous text of a failed reaction. Viewing only the text file, the wrong conclusion about the sequence reaction may be reached. The example in Figure 1 gives an additional indication that more than one product is present. At base 197 the multiple peaks end and a single peak for each base begins. It is at this point that the cloned inserts transition to plasmid sequence.
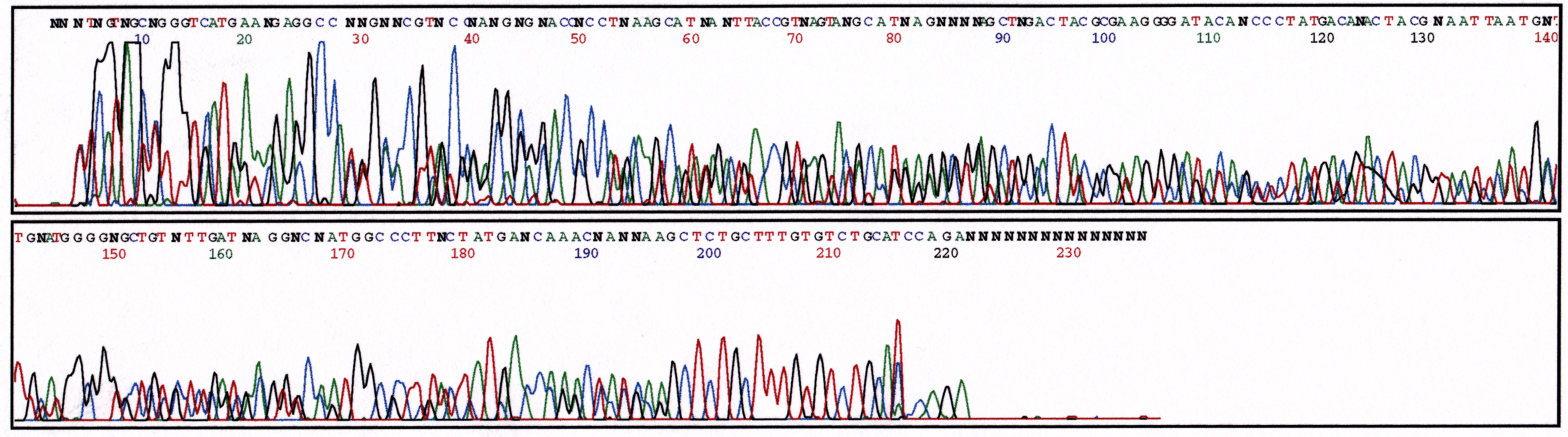
Figure 1. Chromatogram displaying sequence reaction containing two cloned inserts.
The inverse can also occur when sequencing from the plasmid into the insert. Peaks begin as single peaks but transition to multiple peaks often at the restriction site used for cloning the insert.
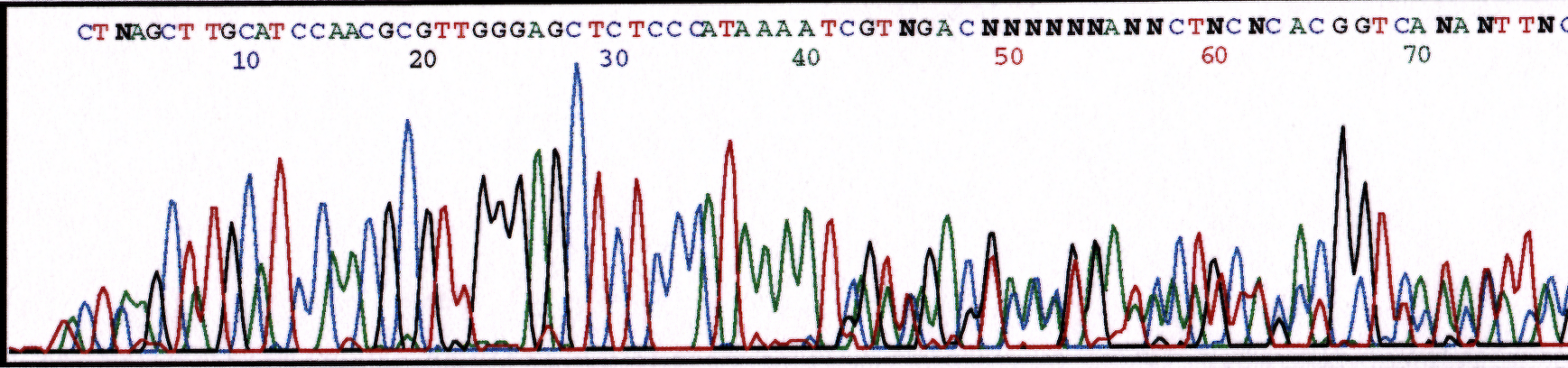
Figure 2. Chromatogram displaying sequence reaction containing two cloned inserts.
Obtaining multiple sequence data in a single sequence reaction is contributed to a variety of problems. It is essential to remember that a sequence reaction uses only one primer. Two primers will result in multiple sequence data. Multiple templates in a submitted sample will likewise result in multiple sequence data. Two clones in a sample can occur when two colonies are inadvertently picked from an agarose plate. Contamination of a glycerol stock or growth media will yield similar results. Precautions are necessary in cloning experiments to ensure that a single clone is present. A restriction digest of the plasmid with insert is recommended to determine the presence of one insert. When sequencing from PCR products it is wise to confirm with an agarose gel that no spurious products are present. A nested primer, designed to anneal to an internal site on the desired PCR product, can be used in situations where multiple products are unavoidable.
