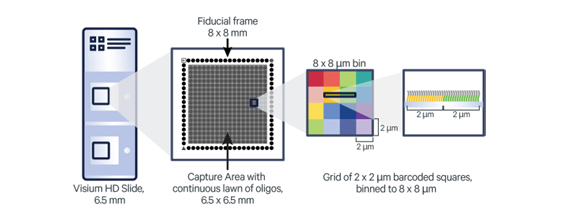
The Visium HD Spatial Gene Expression slide allows for the detection of gene expression across intact fresh frozen tissue. Superimposing the gene expression information upon an image of the tissue allows researchers to understand the spatial organization of cell types.
Visium HD Spatial services are provided in collaboration with the Advanced Light Microscopy Core and the Bioinformatics and Analytics Core.
Additional documentation and resources are available on the 10x Genomics support site
A meeting is required with core staff to initiate a Visium spatial project. The meeting will discuss experimental goals as well as project turnaround times, tissue processing, and sequencing requirements. Arrangements for a meeting to discuss a project can be made by sending an email to Nathan Bivens, Genomics Core director.
All projects must be initiated with a quote, and quote accepted, before tissue processing can be scheduled. .
The 10x Spatial Calculator may be used to estimate the amount of sequence required for a project.
The initial step in a Visium Spatial protocol is the cryosectioning and placement of the tissue on the Visium slide. The 10x Genomics Tissue Preparation Guide provides guidance on the handling of fresh frozen (FF) tissue samples and generating tissue slides. Protocol outlines fixation, H&E staining, and imaging.
Support and services for tissue section, staining and imaging is available through the Advanced Light Microscopy Core.
| Sequence Depth/Coverage | Read Type | Index Type | Read Length | ||
| 550 million read pairs per capture area | Paired end | unique dual-index | Read 1: 43 bases Read 2: 75 bases | ||
